日本語
English
Enzyme Kinetics Module 解析手法
| 基質なし | pH 依存性(6種)、初速度、熱変性、変性剤変性、指数減衰(4種)、直線 |
|---|---|
| 基質1個 | 基質1(6種)、基質1 – 阻害剤1(8種)、基質1 – 強結合阻害剤(4種)、酵素活性剤あり(Nonessential・SA Complex ) |
| 基質2個 | 基質2(3種) |
- 組み込まれている各基質数での実験のタイプと式およびグラフの種類
■ 基質の数 = None
-
- pH Rate Profile
- Equations
- Single pKa (Rising)
Vmaxlim/(1+10^(pKa-pH)) - Single pKa (Falling)
Vmaxlim/(1+10^(pH-pKa)) - Single pKa With Offset
(Vlim1 +Vlim2*10^(pH-pKa))/(1+10^(pH-pKa)) - Bell Shaped
(Vmaxlim/(1+10^(pKa-pH) + 10^(pH-pKb)) - Bell Shaped With Plateau
Vmaxlim*alpha+10^(pKb-pH))/1+ 10^(pKa+pKb-2*pH)+10^(pKb-pH)+10^(pH-pKc)) - Plateau Shaped
(Vmaxpla+Vmaxlim*10^(pH-pKb))/1+ 10^(pKa-pH)+10^(pH-pKb))
- Single pKa (Rising)
- Graphs
- y vs. pH
- log(y) vs.pH
- Residual Graphs
- Equations
- pH Rate Profile
-
- Exponential Decay
- Equations
- Single Exponential
ymax*exp(-k*t) - Single Exponential + Offset
y0+ymax*exp(-k*t) - Double Exponential
ymax1*exp(-k1*t)+ymax2*exp(-k2*t) - Double Exponential + Offset
y0+ymax1*exp(-k1*t)+ymax2*exp(-k2*t)
- Single Exponential
- Graphs
- y(residuals) vs. time
- log(y) vs. t
- Residual Graphs
- Equations
- Regression
- Equations
- Linear
y0+a*x
- Linear
- Graphs
- y vs. x
- Residual Graphs
- Equations
- Exponential Decay
■ 基質数 = 1
-
- Single Substrate
- Equations
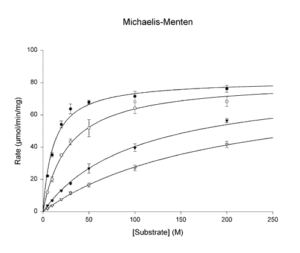
- Michaelis-Menten
v = Vmax*S/(Km+S) - Substrate Inhibition (Uncompetitive)
y= Vmax/(1+Km/S+S/Ki) - Substrate Activation (Ordered)
v = Vmax*(S/Ks)2/alpha/(1+S/Ks+(S/Ks)2/alpha) - Substrate Activation (Random)
v = Vmax*(S/Ks)2/alpha/(1+S/Ks+S/(Ks/alpha)+(S/Ks)2/alpha) - Hill
v = Vmax*Sn/(Kmn+Sn) - Isoenzyme
v = Vmax1*(S/Km1)/(1+S/Km1)+Vmax2*(S/Km2)/(1+S/Km2)
- Michaelis-Menten
- Graphs
- Equations
- Single Substrate
-
- Single Substrate – Single Inhibitor
- Equations
- Competitive (Full)
v = Vmax/(1+(Km/S)*(1+I/Ki)) - Competitive (Partial)
v = Vmax/(1+(Km/S)*(1+I/Ki1)/(1+I/Ki2)) - Noncompetitive (Full)
v = Vmax/((1+I/Ki)*(1+Km/S)) - Noncompetitive (Partial)
v = Vmax/((1+Km/S)*(1+I/Ki)/(1+I*beta/Ki)) - Mixed (Full)
v = Vmax/((Km/S)*(1+I/Ki)+(1+I/(alpha*Ki))) - Mixed (Partial)
v = Vmax*((1+beta*I/(alpha*Ki))/(1+I/(alpha*Ki)))/ (1+(Km/S)*(1+I/Ki)/(1+I/(alpha*Ki))) - Uncompetitive (Full)
v = Vmax/(1+I/Ki+Km/S) - Uncompetitive (Partial)
v = Vmax*(1+beta*(I/Ki))/(1+I/Ki+Km/S)
- Competitive (Full)
- Graphs
- Equations
- Single Substrate – Single Inhibitor
-
- Tight Binding Inhibitor
- Equations
- Competitive Tight
- v0 = Vmax/(Km/S+1)
- Kap = Ki*((S/Km)+1)
- v = (v0/(2*E))*(E-I-Kap+sqrt((E-I-Kap)2+4*E*Kap))
- Noncompetitive Tight
- v0 = Vmax/(Km/S+1)
- v = (v0/(2*E))*(E-I-Ki+sqrt((E-I-Ki)2+4*E*Ki))
- Uncompetitive Tight
- v0 = Vmax/(Km/S+1)
- Kap = Ki*((Km/S)+1)
- v = (v0/(2*E))*(E-I-Kap+sqrt((E-I-Kap)2+4*E*Kap))
- Mixed Tight
- E = 27
- alpha = 5.6
- v0 = Vmax/(Km/S+1)
- Iap = Ki*((S/Km)+1)*alpha/(alpha+S/Km)
- v = ((v0-vinf)/(2*E))*(((v0+vinf)/(v0-vinf))*E-I-Iap+sqrt((E-I-Iap)2+4*E*Iap))
- Competitive Tight
- Graphs
- Michaelis-Menten
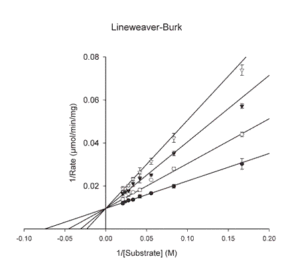
- Lineweaver-Burk
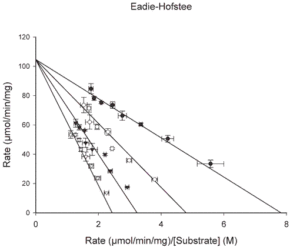
- Eadie-Hofstee
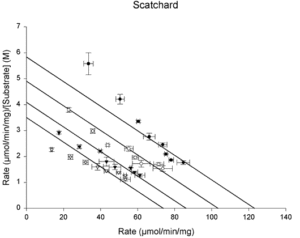
- Scatchard
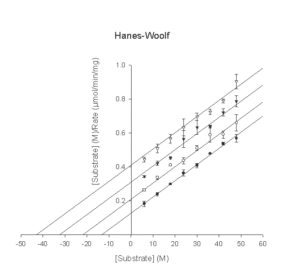
- Hanes-Woolf
- Hill
- Dixon
- Residual Graphs
- Equations
- Enzyme Activator – Nonessential
- Equations
- Nonessential
v = Vmax*(1+beta*A/(alpha*Ka))/((Km/S)*(1+A/Ka)+(1+A/(alpha*Ka)))
- Nonessential
- Graphs
- Michaelis-Menten
- Lineweaver-Burk
- Eadie-Hofstee
- Scatchard
- Hanes-Woolf
- Hill
- Residual Graphs
- Equations
- Enzyme Activator – SA Complex
- Equations
- Specific
v = Vmax*S/Km/(1+S/Km+Ka/A) - Catalytic
v = Vmax*S/Km/(1+Ka/A+S/Km*(1+Ka/A)) - Mixed
v = Vmax*S/Km/(1+Kas/A+S/Km*(1+Kac/A))
- Specific
- Graphs
- Michaelis-Menten
- Lineweaver-Burk
- Eadie-Hofstee
- Scatchard
- Hanes-Woolf
- Hill
- Residual Graphs
- Equations
- Tight Binding Inhibitor
■ 基質数 = 2
-
- Two Substrate
- Equations
- Random Bi-Bi (Sequential)
y = Vmax*A*B/(alpha*Ka*Kb+Kb*A+Ka*B)+A*B) - Ordered Bi-Bi (Sequential)
y = Vmax*A*B/(Ka*Kb+Kb*A+A*B) - Ping Pong Bi-Bi (Nonsequential)
if ((A=0)(B=0), 0, y = Vmax*A*B/(Kb*A+Ka*B+A*B))
- Random Bi-Bi (Sequential)
- Graphs
- Michaelis-Menten A
- Michaelis-Menten B
- Lineweaver-Burk A
- Lineweaver-Burk B
- Eadie-Hofstee A
- Eadie-Hofstee B
- Scatchard A
- Scatchard B
- Hanes-Woolf A
- Hanes-Woolf B
- Hill A
- Hill B
- Residual Graphs
- Equations
- Two Substrate
- ユーザー定義式の登録
- 以下の実験タイプには式を追加することが出来ます。
- pH Rate Profile
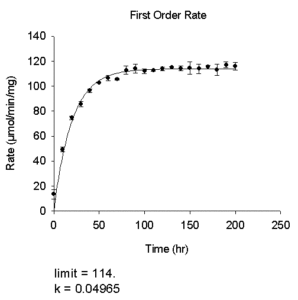
- First Order Rate
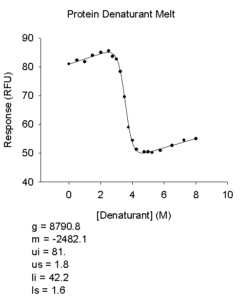
- Protein Denaturant Melt
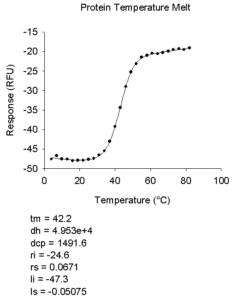
- Protein Temperature Melt
- Exponential Decay
- Regression
- 以下の実験タイプには式を追加することが出来ます。