 |
| サイトマップ | |
|
 |
| サイトマップ | |
|
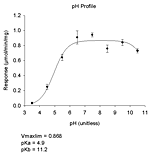
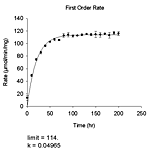
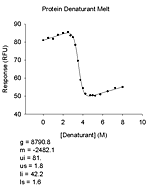
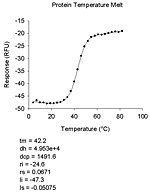
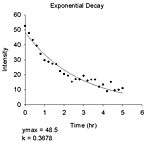
| 基質なし | pH 依存性(6種)、初速度、熱変性、変性剤変性、指数減衰(4種)、直線 |
|---|---|
| 基質1個 | 基質1(6種)、基質1 – 阻害剤1(8種)、基質1 – 強結合阻害剤(4種)、酵素活性剤あり(Nonessential・SA Complex ) |
| 基質2個 | 基質2(3種) |
■ 基質の数 = None
■ 基質数 = 1
■ 基質数 = 2